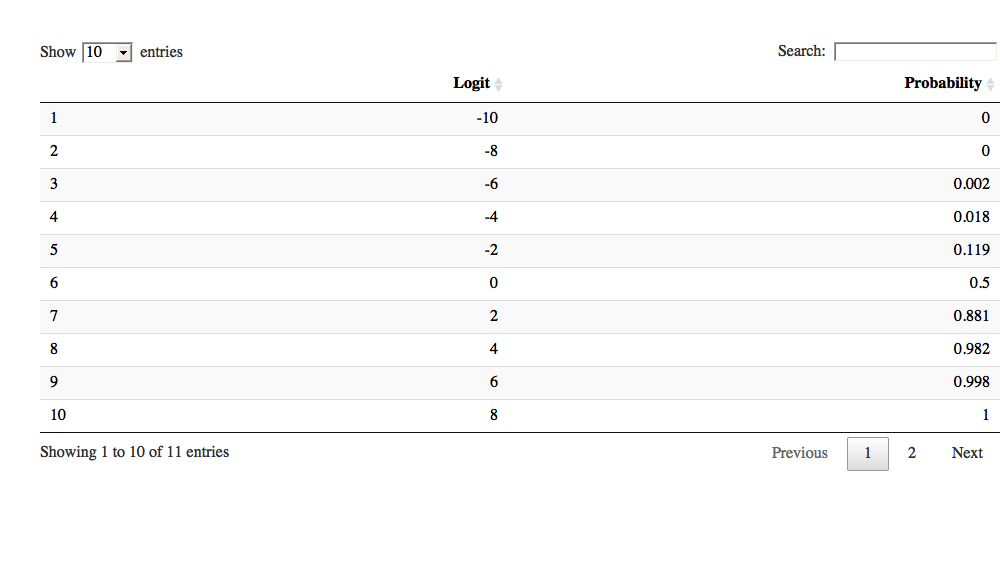
Bei der Textanalyse (Textmining) ist die Sentiment-Analyse eine typische Tätigkeit. Natürlich steht und fällt die Qualität der Sentiment-Analyse mit der Qualität des verwendeten Wörterbuchs (was nicht heißt, dass man nicht auch auf andere Klippen schellen kann).
Der Zweck dieses Posts ist es, eine Sentiment-Lexikon in deutscher Sprache einzulesen.
Dazu wird das Sentiment-Lexikon dieser Quelle verwendet (CC-BY-NC-SA 3.0). In diesem Paper finden sich Hintergründe. Von dort lassen sich die Daten herunter laden. Im folgenden gehe ich davon aus, dass die Daten herunter geladen sind und sich im Working Directory befinden.
Wir benötigen diese Pakete (es ginge auch über base):
library(stringr)
library(readr)
library(dplyr)
Dann lesen wir die Daten ein, zuerst die Datei mit den negativen Konnotationen:
neg_df <- read_tsv("SentiWS_v1.8c_Negative.txt", col_names = FALSE)
names(neg_df) <- c("Wort_POS", "Wert", "Inflektionen")
glimpse(neg_df)
#> Observations: 1,818
#> Variables: 3
#> $ Wort_POS <chr> "Abbau|NN", "Abbruch|NN", "Abdankung|NN", "Abdämp...
#> $ Wert <dbl> -0.0580, -0.0048, -0.0048, -0.0048, -0.0048, -0.3...
#> $ Inflektionen <chr> "Abbaus,Abbaues,Abbauen,Abbaue", "Abbruches,Abbrü...
Dann parsen wir aus der ersten Spalte (Wort_POS) zum einen den entsprechenden Begriff (z.B. “Abbau”) und zum anderen die Wortarten-Tags (eine Erläuterung zu den Wortarten-Tags findet sich hier).
neg_df %>%
mutate(Wort = str_sub(Wort_POS, 1, regexpr("\\|", .$Wort_POS)-1),
POS = str_sub(Wort_POS, start = regexpr("\\|", .$Wort_POS)+1)) -> neg_df
str_sub parst zuerst das Wort. Dazu nehmen wir den Wort-Vektor Wort_POS, und für jedes Element wird der Text von Position 1 bis vor dem Zeichen | geparst; da der Querstrich ein Steuerzeichen in Regex muss er escaped werden. Für POS passiert das gleiche von Position |+1 bis zum Ende des Text-Elements.
Das gleiche wiederholen wir für positiv konnotierte Wörter.
pos_df <- read_tsv("SentiWS_v1.8c_Positive.txt", col_names = FALSE)
names(pos_df) <- c("Wort_POS", "Wert", "Inflektionen")
pos_df %>%
mutate(Wort = str_sub(Wort_POS, 1, regexpr("\\|", .$Wort_POS)-1),
POS = str_sub(Wort_POS, start = regexpr("\\|", .$Wort_POS)+1)) -> pos_df
Schließlich schweißen wir beide Dataframes in einen:
bind_rows("neg" = neg_df, "pos" = pos_df, .id = "neg_pos") -> sentiment_df
sentiment_df %>% select(neg_pos, Wort, Wert, Inflektionen, -Wort_POS) -> sentiment_df
knitr::kable(head(sentiment_df))
| neg_pos | Wort | Wert | Inflektionen |
|---|---|---|---|
| neg | Abbau | -0.058 | Abbaus,Abbaues,Abbauen,Abbaue |
| neg | Abbruch | -0.005 | Abbruches,Abbrüche,Abbruchs,Abbrüchen |
| neg | Abdankung | -0.005 | Abdankungen |
| neg | Abdämpfung | -0.005 | Abdämpfungen |
| neg | Abfall | -0.005 | Abfalles,Abfälle,Abfalls,Abfällen |
| neg | Abfuhr | -0.337 | Abfuhren |
Literatur
R. Remus, U. Quasthoff & G. Heyer: SentiWS - a Publicly Available German-language Resource for Sentiment Analysis. In: Proceedings of the 7th International Language Ressources and Evaluation (LREC’10), 2010