reading time: 15-20 min.
Slidify is a cool tool to render HTML5 slide decks, see here, here or here for examples.
Features include:
- reproducibility. You write your slide deck as you would write any other text, similar to Latex/Beamer. But you write using Markdown, which is easier and less clumsy. As you write plain text, you are free to use git.
- modern look. Just a website, nothing more. But with cool, modern features.
- techiwecki. Well, probably techie-minded persons will love that more than non-techies… Check this tutorial out as a starter.
Basically, slidify procudes a website:
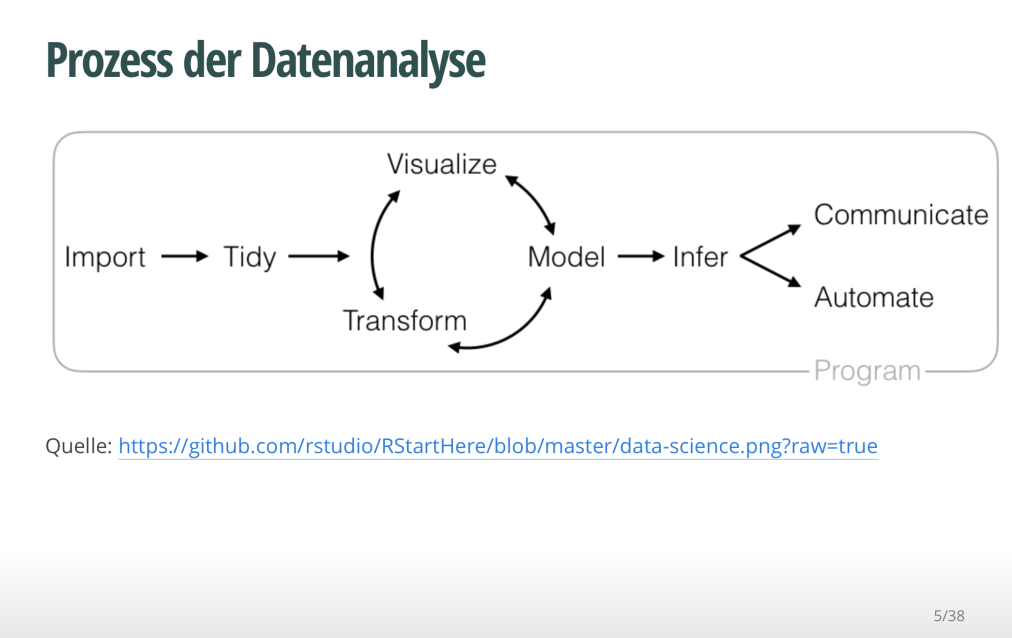
While it is quite straight forward to write up your slidify presentations, some customization is a bit more of a hassle.
Let’s assume you would like to include a logo to your presentation. How to do that?
Logo page
There is an easy solution: In your slidify main file (probably something like index.Rmd) you will find the YAML header right at the start of the file:
---
title : "Test with Logo"
author : "Dr. Zack"
widgets : [mathjax, quiz, bootstrap, interactive] # {mathjax, quiz, bootstrap}
ext_widgets : {rCharts: [libraries/nvd3, libraries/leaflet, libraries/dygraphs]}
mode : selfcontained # {standalone, draft}
logo : logo.png
biglogo : logo.png
---
To get your logo page, do the following two steps:
- In your project folder, go to assets/img. Drop your logo file there (png and jpg will both work)
- Then adjust your YAML header by adding two lines:
logo : logo.png
biglogo : logo.png
Done!
Two changes will now take place. First, you will have a logo page, where nothing but your logo will show up (biglogo.png):

Second, a following page, the title page, will also show you logo (logo.png), but smaller and with a little animation (in the default):

Note that there are a number of other variables that you can define in the YAML header.
Background image on title page
Now, a little more fancy. What about a cool background image on your first page? OK, let’s check it out. This will be the output:

So what did I do?
I defined a CSS class as follows:
.title-slide {
background-image: url(https://goo.gl/gAeQqj);
}
The picture is from WikiMedia Commons, published in the public domain.
Then, I saved this code as a css file (name does not matter; in my case title_slide_bg.css) in this folder:
[project_folder]/assets/css.
That’s it. Wait, don’t forget to slidify("my_deck").
You will say, that’s ok, but I want a footer or a header line, because that’s where I like to put my logo (accustomed to…).
Footer/header with logo
The solution I used (there are surely a number of different) consisted of rewriting/customizing the template of the standard slide slide.html, adding the footer/header with logo.
So, the basic slide template looks like this:
<slide class="" id="" style="background:};">
<hgroup>
}
</hgroup>
<article data-timings="">
}
</article>
<!-- Presenter Notes -->
<aside class="note" id="">
<section>
}
</section>
</aside>
</slide>
I added some lines to add an object (logo) at a certain position; so my slide.html file looked like this:
<slide class="" id="">
<hgroup>
}
</hgroup>
<article>
}
<footer class = 'logo'>
<div style="position: absolute; left: 1000px; top: 50px; z-index:100">
<img src = "assets/img/logo.png" height="80" width="80">
</div>
</footer>
</article>
</slide>
Now, we have to save this file under [project folder]/assets/layouts.
The name does not matter; any html-file in this folder will be parsed by slidify. Here come the header with logo:

You can adapt size and position of the logo with the img html function.
That’s it! Happy slidifying!
You will find the code on this github repo, along with the slidify-presentation.