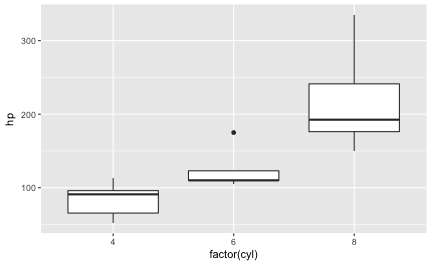
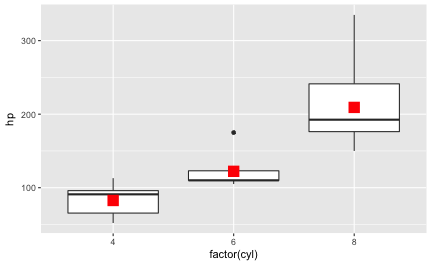
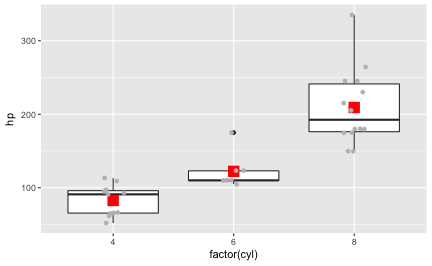
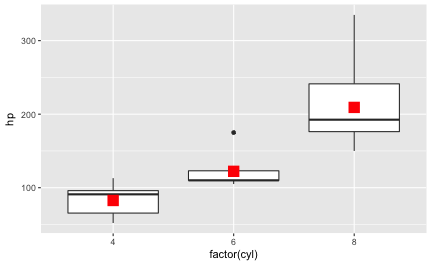
This post presents a compilation of links to psychology papers; I have chosen papers I find intriguing particularly for working in class. All papers are open access (or a from open access repositories) which renders classroom work easier. The papers are collected from a broad range of topics but mostly with focus on general interest. The perspective is an applied one; I have not tried to select based on methodological rigor. The collection is structured along the well-known classification of psychological work: social, personality, cognitive. I have added ‘social media/ psychoinformatics’ as this reflects a topic I am quite interested in.
I am unsure about compilations of ‘must read’ psych articles, but I have found some. Such sites may provide a more succinct and broader perspective on much read or influential or interesting or high-qualitative science papers. Here’s a short list:
- Altmetric Top 100 - 2016 (filter on ‘studies in human society’)
- Altmetric Top 100 - 2015 (filter on ‘studies in human society’)
- Frontiers in Psychology (click on ‘most cited’)
Social psychology
- Meta-Milgram: An Empirical Synthesis of the Obedience Experiments
- Behavioral Priming: It’s All in the Mind, but Whose Mind?
- Always Gamble on an Empty Stomach: Hunger Is Associated with Advantageous Decision Making
- A Virtual Reprise of the Stanley Milgram Obedience Experiments
- The Power of Kawaii: Viewing Cute Images Promotes a Careful Behavior and Narrows Attentional Focus
- Non-Disruptive Tactics of Suppression Are Superior in Countering Terrorism, Insurgency, and Financial Panics
Personality/ Individual differences
- Loss of Control Increases Belief in Precognition and Belief in Precognition Increases Control Temptation at Work
- The Distance Between Mars and Venus: Measuring Global Sex Differences in Personality
- Personality, Gender, and Age in the Language of Social Media: The Open-Vocabulary Approach
- A Propaganda Index for Reviewing Problem Framing in Articles and Manuscripts: An Exploratory Study
- Are women better than men at multi-tasking?
- Business culture and dishonesty in the banking industry
- The Negative Association between Religiousness and Children’s Altruism across the World
- Does happiness itself directly affect mortality? The prospective UK Million Women Study
Cognitive psychology
- The Eyes Don’t Have It: Lie Detection and Neuro-Linguistic Programming
- Human language reveals a universal positivity bias