I found myself doing the following: I had a bunch of predictors, one (numeric) outcome, and wanted to run I simple regression for each of the predictors. Having a bunch of model results, I would like to have them bundled in one data frame.
So, here is one way to do it.
First, load some data.
data(mtcars)
str(mtcars)
## 'data.frame': 32 obs. of 11 variables:
## $ mpg : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
## $ cyl : num 6 6 4 6 8 6 8 4 4 6 ...
## $ disp: num 160 160 108 258 360 ...
## $ hp : num 110 110 93 110 175 105 245 62 95 123 ...
## $ drat: num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
## $ wt : num 2.62 2.88 2.32 3.21 3.44 ...
## $ qsec: num 16.5 17 18.6 19.4 17 ...
## $ vs : num 0 0 1 1 0 1 0 1 1 1 ...
## $ am : num 1 1 1 0 0 0 0 0 0 0 ...
## $ gear: num 4 4 4 3 3 3 3 4 4 4 ...
## $ carb: num 4 4 1 1 2 1 4 2 2 4 ...
Say, mpg is our outcome/ criterion. The rest of the variables are predictors.
Then, some packages.
library(dplyr)
library(purrr)
library(tibble)
library(ggplot2)
library(stringr)
library(tidyr)
library(broom)
library(scales)
For illustration, let’s run a regression with each and all of the predictors as a preliminary step.
lm(mpg ~ ., data = mtcars) %>% glance
## r.squared adj.r.squared sigma statistic p.value df logLik
## 1 0.8690158 0.8066423 2.650197 13.93246 3.793152e-07 11 -69.85491
## AIC BIC deviance df.residual
## 1 163.7098 181.2986 147.4944 21
lm(mpg ~ ., data = mtcars) %>% summary %>% tidy
## term estimate std.error statistic p.value
## 1 (Intercept) 12.30337416 18.71788443 0.6573058 0.51812440
## 2 cyl -0.11144048 1.04502336 -0.1066392 0.91608738
## 3 disp 0.01333524 0.01785750 0.7467585 0.46348865
## 4 hp -0.02148212 0.02176858 -0.9868407 0.33495531
## 5 drat 0.78711097 1.63537307 0.4813036 0.63527790
## 6 wt -3.71530393 1.89441430 -1.9611887 0.06325215
## 7 qsec 0.82104075 0.73084480 1.1234133 0.27394127
## 8 vs 0.31776281 2.10450861 0.1509915 0.88142347
## 9 am 2.52022689 2.05665055 1.2254035 0.23398971
## 10 gear 0.65541302 1.49325996 0.4389142 0.66520643
## 11 carb -0.19941925 0.82875250 -0.2406258 0.81217871
Of interest, no p-value is below .05.
Now come the main part, let’s run multiple regression and then combine the results.
mtcars %>%
dplyr::select(-mpg) %>%
map(~lm(mtcars$mpg ~ .x, data = mtcars)) %>%
map(summary) %>%
map("coefficients") %>%
do.call(rbind.data.frame, .) %>%
rownames_to_column %>%
as_tibble %>%
setNames(c("predictor", "b", "SE", "t", "p")) %>%
dplyr::arrange(p) %>%
dplyr::filter(!str_detect(predictor, "(Intercept)")) %>%
mutate(predictor = str_sub(predictor, start = 1, end = str_length(predictor)-3))
## # A tibble: 10 × 5
## predictor b SE t p
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 wt -5.34447157 0.559101045 -9.559044 1.293959e-10
## 2 cyl -2.87579014 0.322408883 -8.919699 6.112687e-10
## 3 disp -0.04121512 0.004711833 -8.747152 9.380327e-10
## 4 hp -0.06822828 0.010119304 -6.742389 1.787835e-07
## 5 drat 7.67823260 1.506705108 5.096042 1.776240e-05
## 6 vs 7.94047619 1.632370025 4.864385 3.415937e-05
## 7 am 7.24493927 1.764421632 4.106127 2.850207e-04
## 8 carb -2.05571870 0.568545640 -3.615750 1.084446e-03
## 9 gear 3.92333333 1.308130699 2.999191 5.400948e-03
## 10 qsec 1.41212484 0.559210130 2.525213 1.708199e-02
Some explanation:
- l2: deselect the outcome variable, so that we can address “all” variables in the next lines
- l3: map each list element to
lm;.xis a placeholder for all the list elements (here predictors) - l4: now get the summary of each lm. More specifically, we have a number of lm models, which are stored as list elements. Now we apply the
summaryfunction to each of those list elements (lm results). - l5: address (extract) the
coefficientssubelement from each list element - l6: rownames should be their own column
- l7: tibbles are nic
- l8: make the col names typing-friendly
- l9: filter those lines, where
predictoris not equal to “(Intercept)”. - l10: change the values of
predictorsuch that the strange end part “..x” is removed
Puh, quite some hassle.
Now, for completeness, let’s look at the $R^2$.
mtcars %>%
dplyr::select(-mpg) %>%
map(~lm(mtcars$mpg ~ .x, data = mtcars)) %>%
map(glance) %>%
do.call(rbind.data.frame, .) %>%
rownames_to_column %>%
as_tibble
## # A tibble: 10 × 12
## rowname r.squared adj.r.squared sigma statistic p.value df
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
## 1 cyl 0.7261800 0.7170527 3.205902 79.561028 6.112687e-10 2
## 2 disp 0.7183433 0.7089548 3.251454 76.512660 9.380327e-10 2
## 3 hp 0.6024373 0.5891853 3.862962 45.459803 1.787835e-07 2
## 4 drat 0.4639952 0.4461283 4.485409 25.969645 1.776240e-05 2
## 5 wt 0.7528328 0.7445939 3.045882 91.375325 1.293959e-10 2
## 6 qsec 0.1752963 0.1478062 5.563738 6.376702 1.708199e-02 2
## 7 vs 0.4409477 0.4223126 4.580827 23.662241 3.415937e-05 2
## 8 am 0.3597989 0.3384589 4.902029 16.860279 2.850207e-04 2
## 9 gear 0.2306734 0.2050292 5.373695 8.995144 5.400948e-03 2
## 10 carb 0.3035184 0.2803024 5.112961 13.073646 1.084446e-03 2
## # ... with 5 more variables: logLik <dbl>, AIC <dbl>, BIC <dbl>,
## # deviance <dbl>, df.residual <int>
Ok, I admit, I like plotting, particularly with ggplot2.
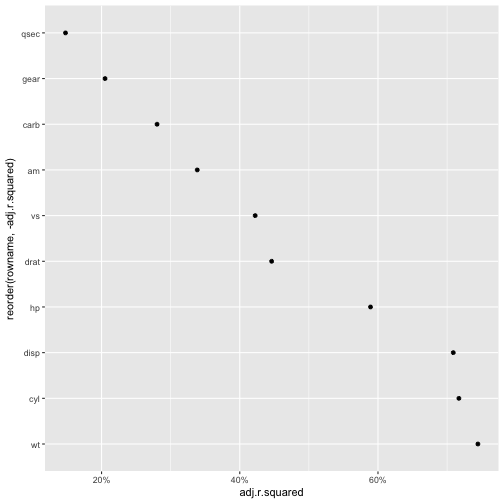
First, the $R^2$ for all models:
mtcars %>%
dplyr::select(-mpg) %>%
map(~lm(mtcars$mpg ~ .x, data = mtcars)) %>%
map(glance) %>%
do.call(rbind.data.frame, .) %>%
rownames_to_column %>%
as_tibble %>%
ggplot(aes(x = reorder(rowname, -adj.r.squared), y = adj.r.squared)) +
geom_point() +
coord_flip() +
scale_y_continuous(labels = scales::percent)
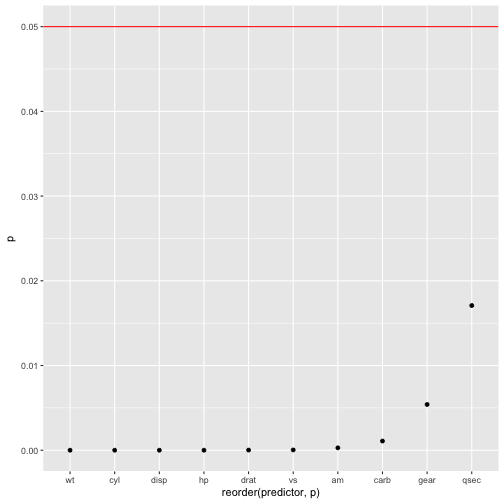
Last, the p-values of each predictor (OMG).
mtcars %>%
dplyr::select(-mpg) %>%
map(~lm(mtcars$mpg ~ .x, data = mtcars)) %>%
map(summary) %>%
map("coefficients") %>%
do.call(rbind.data.frame, .) %>%
rownames_to_column %>%
as_tibble %>%
setNames(c("predictor", "b", "SE", "t", "p")) %>%
dplyr::arrange(p) %>%
dplyr::filter(!str_detect(predictor, "(Intercept)")) %>%
mutate(predictor = str_sub(predictor, start = 1, end = str_length(predictor)-3)) %>%
ggplot(aes(x = reorder(predictor,p), y = p)) +
geom_point() +
geom_hline(yintercept = .05, color = "red")