library(tidyverse)
d_path <- "https://vincentarelbundock.github.io/Rdatasets/csv/palmerpenguins/penguins.csv"
d <- read_csv(d_path)
nrow(d)[1] 344May 14, 2023
Liefern Sie einen visuellen Überblick über fehlende Werte im Datensatz penguins!
d_na_only <-
d %>%
rowwise() %>%
mutate(na_n = sum(is.na(cur_data()))) %>%
ungroup()
d_na_only %>%
ggplot(aes(x = na_n)) +
geom_bar()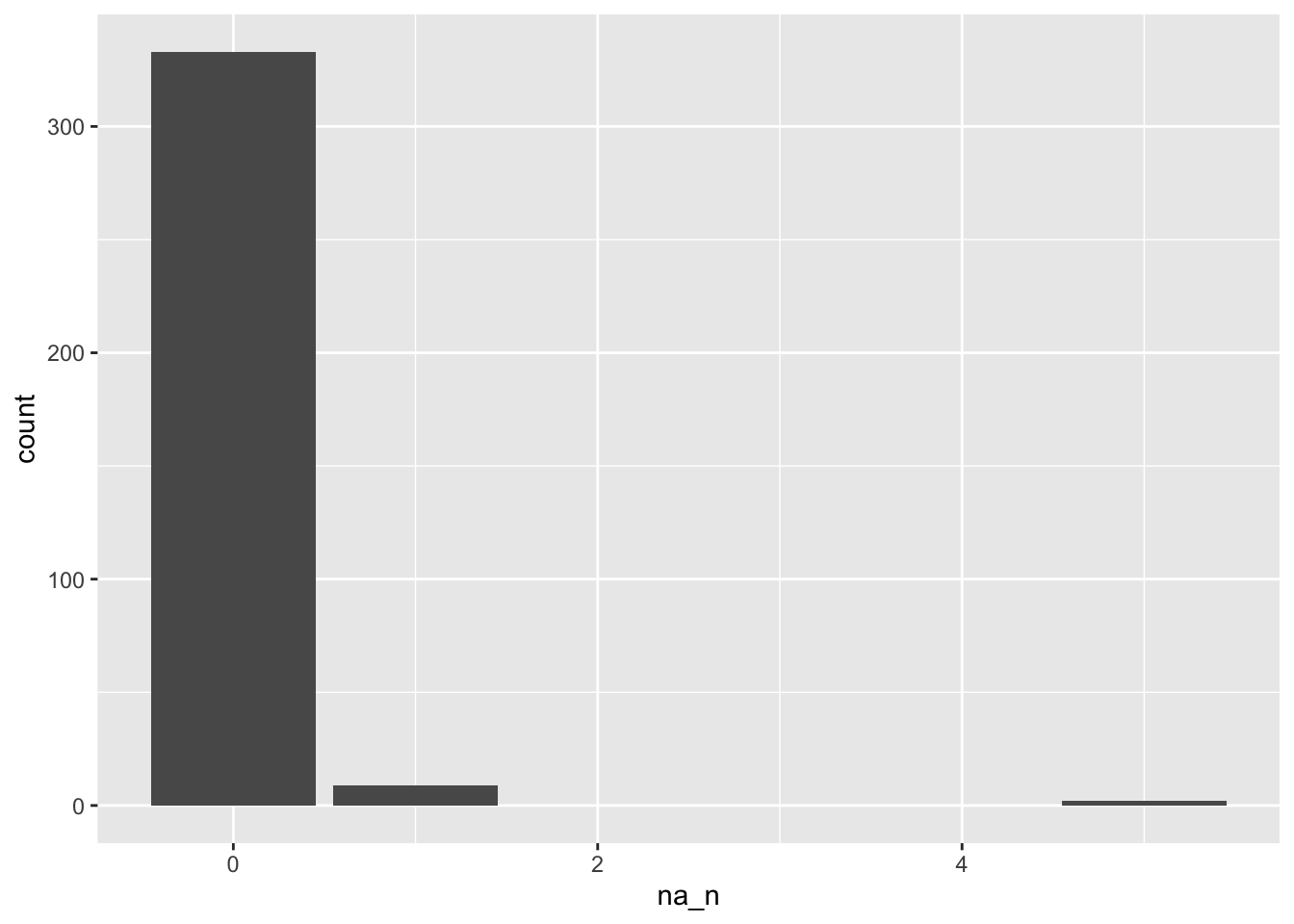
Categories:
---
exname: filter-na4
expoints: 1
extype: string
exsolution: NA
categories:
- 2023
- eda
- na
- string
date: '2023-05-14'
slug: filter-na4
title: filter-na4
---
# Aufgabe
Liefern Sie einen visuellen Überblick über fehlende Werte im Datensatz `penguins`!
</br>
</br>
</br>
</br>
</br>
</br>
</br>
</br>
</br>
</br>
# Lösung
## Setup
```{r}
library(tidyverse)
d_path <- "https://vincentarelbundock.github.io/Rdatasets/csv/palmerpenguins/penguins.csv"
d <- read_csv(d_path)
nrow(d)
```
## Weg 1
```{r}
library(visdat)
vis_dat(d)
```
## Weg 2
```{r}
d_na_only <-
d %>%
rowwise() %>%
mutate(na_n = sum(is.na(cur_data()))) %>%
ungroup()
d_na_only %>%
ggplot(aes(x = na_n)) +
geom_bar()
```
---
Categories:
- 2023
- eda
- na
- string