April 28, 2023
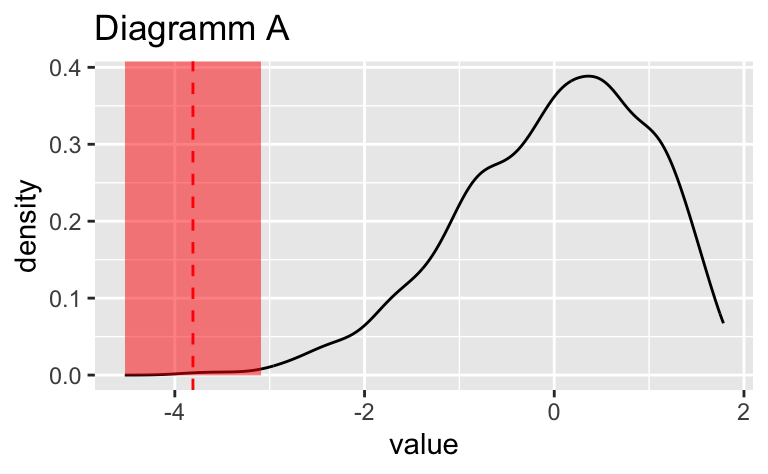
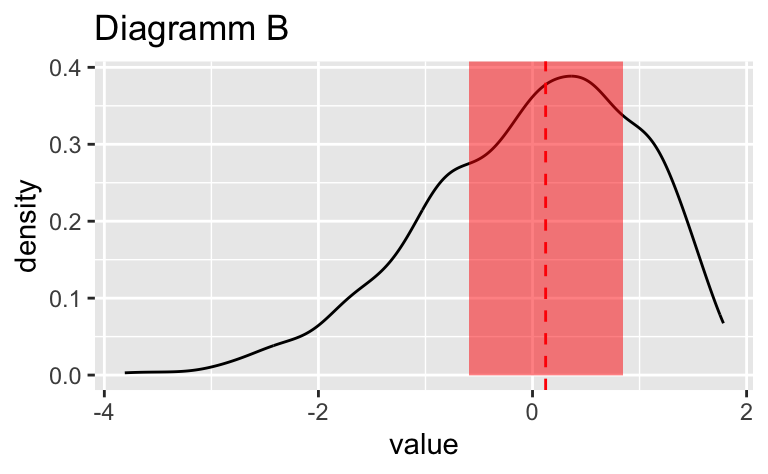
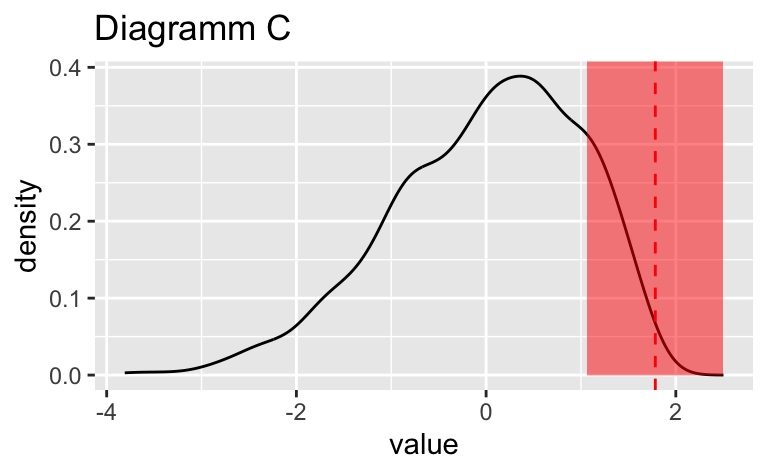
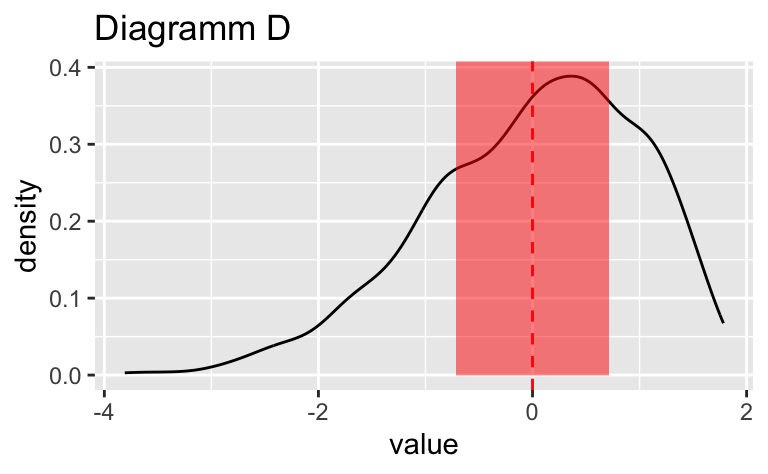
Wählen Sie das Diagramm, in dem der vertikale gestrichelte Linie am genauesten die Position des Medians (\(Md\)) widerspiegelt.




Das Diagramm B zeigt den Median am genauesten.
Categories:
---
exname: Streuung-Histogramm
extype: schoice
exsolution: r mchoice2string(sol_df$is_correct, single = TRUE)
exshuffle: no
categories:
- eda
- variability
- dyn
- schoice
date: '2023-04-28'
slug: Streuung-Histogramm
title: Streuung-Histogramm
---
```{r libs, include = FALSE}
library(tidyverse)
library(mosaic)
#library(testthat)
library(e1071) # for skewness
library(glue)
library(exams)
```
```{r global-knitr-options, include=FALSE}
knitr::opts_chunk$set(fig.pos = 'H',
fig.asp = 0.618,
fig.width = 4,
fig.cap = "",
fig.path = "",
cache = FALSE)
```
# Aufgabe
```{r solution, echo = FALSE}
sol_set <- c(
#"symm",
# "uniform",
"linksschief1", "linksschief2",
"rechtsschief1", "rechtsschief2"
# ,"rechtsschief3"
)
sol <- sample(sol_set, 1)
```
Wählen Sie das Diagramm, in dem der vertikale gestrichelte Linie am genauesten die Position des Medians ($Md$) widerspiegelt.
```{r generate-data, echo = FALSE}
n <- 1000
d_symm <- rnorm(n = n) %>% zscore() %>% as_tibble()
d_left1 <- rbeta(n, 5, 2) %>% zscore() %>% as_tibble()
d_right1 <- rlnorm(n) %>% zscore() %>% as_tibble()
d_right2 <- rlnorm(n) %>% zscore()%>% as_tibble()
d_left2 <- -rexp(n, 10) %>% zscore()%>% as_tibble()
d_uniform <- runif(n = n) %>% zscore() %>% as_tibble()
# heavy (right) tailed pareto distribution:
qpareto <- function(u, a=0.8, b=1) b/(1-u)^(1/a)
rpareto <- function(n, a=0.8, b=1) qpareto(runif(n),a,b)
d_right3 <- rpareto(n) %>% zscore() %>% as_tibble()
d_raw <- tibble(
data = list(
#d_symm,
#d_uniform,
d_left1,
d_left2,
d_right1,
d_right2
#,d_right3
)) %>%
mutate(id = sol_set)
d <-
d_raw %>%
mutate(quantiles = map(data, ~ quantile(.x[[1]]))) %>%
mutate(skew = map_dbl(data, ~ skewness(.x[[1]]))) %>%
mutate(avg = map_dbl(data, ~ mean(.x[[1]]))) %>%
mutate(iqr = map_dbl(data, ~ IQR(.x[[1]]))) %>%
unnest_wider(quantiles) %>%
sample_n(size = nrow(.)) # shuffle it
d_selected_distribution <-
d %>%
filter(id == sol)
d_selected_long <-
d_selected_distribution %>%
select(-c(id, skew, `75%`, `25%`)) %>%
pivot_longer(-c(data, iqr)) %>%
mutate(plot_id = LETTERS[1:nrow(.)])
```
```{r generate-histograms, echo = FALSE, message = FALSE, fig.show='hold', comment="", results="hide", warning=FALSE}
gg_hist <- function(data, plot_id, value, iqr) {
ggplot(data = data) +
aes(x = value) +
geom_density() +
ggtitle(glue("Diagramm {plot_id}")) +
annotate("rect", xmin = value - iqr/2, xmax = value + iqr/2, ymin = 0, max = Inf, fill = "red", alpha = .5) +
geom_vline(xintercept = value, linetype = "dashed", color = "red")
}
d_selected_long %>%
select(-name) %>%
pmap(gg_hist)
```
```{r compute-sol-df, echo = FALSE}
sol_df <-
d_selected_long %>%
mutate(is_correct = name == "50%")
```
```{r questionlist, echo = FALSE, results = "asis"}
exams::answerlist(sol_df$plot_id, markup = "markdown")
```
</br>
</br>
</br>
</br>
</br>
</br>
</br>
</br>
</br>
</br>
# Lösung
Das Diagramm ``r sol_df$plot_id[sol_df$is_correct == TRUE]`` zeigt den Median am genauesten.
```{r solutionlist, echo = FALSE, results = "asis"}
exams::answerlist(ifelse(sol_df$is_correct, "Wahr", "Falsch"), markup = "markdown")
```
---
Categories:
- eda
- streuungsmaß
- variability
- dyn
- schoice